3 Plots In R 3,9/5 2722 reviews
You learned from the Plot chapter that the plot function is used to plot numbers against each other. A 'scatter plot' is a type of plot used to display the relationship between two numerical variables, and plots one dot for each observation. Ggplot2 is now over 10 years old and is used by hundreds of thousands of people to make millions of plots. That means, by-and-large, ggplot2 itself changes relatively little. When we do make changes, they will be generally to add new functions or arguments rather than changing the behaviour of existing functions, and if we do make changes to.
Generic X-Y Plotting
Generic function for plotting of R objects. For more details about the graphical parameter arguments, see par.
For simple scatter plots, plot.default will be used. However, there are plot methods for many R objects, including functions, data.frames, density objects, etc. Use methods(plot) and the documentation for these.

- Keywords
- hplot
Usage
Arguments
the coordinates of points in the plot. Alternatively, a single plotting structure, function or any R object with a plot method can be provided.
the y coordinates of points in the plot, optional if x is an appropriate structure.
Arguments to be passed to methods, such as graphical parameters (see par). Many methods will accept the following arguments:
typewhat type of plot should be drawn. Possible types are
'p'for points,'l'for lines,'b'for both,'c'for the lines part alone of'b','o'for both ‘overplotted’,'h'for ‘histogram’ like (or ‘high-density’) vertical lines,'s'for stair steps,'S'for other steps, see ‘Details’ below,'n'for no plotting.
types give a warning or an error; using, e.g., type = 'punkte' being equivalent to 
type = 'p' for S compatibility. Note that some methods, e.g.
for S compatibility. Note that some methods, e.g.plot.factor, do not accept this.mainan overall title for the plot: see title.
suba sub title for the plot: see title.
xlaba title for the x axis: see title.
ylaba title for the y axis: see title.
aspthe (y/x) aspect ratio, see plot.window.
Details
The two step types differ in their x-y preference: Going from ((x1,y1)) to ((x2,y2)) with (x1 < x2), type = 's' moves first horizontal, then vertical, whereas type = 'S' moves the other way around.
See Also
plot.default, plot.formula and other methods; points, lines, par. For thousands of points, consider using smoothScatter() instead of plot().
For X-Y-Z plotting see contour, persp and image.
Aliases
- plot
Examples
library(graphics)# NOT RUN {require(stats) # for lowess, rpois, rnormplot(cars)lines(lowess(cars))plot(sin, -pi, 2*pi) # see ?plot.function## Discrete Distribution Plot:plot(table(rpois(100, 5)), type = 'h', col = 'red', lwd = 10, main = 'rpois(100, lambda = 5)')## Simple quantiles/ECDF, see ecdf() {library(stats)} for a better one:plot(x <- sort(rnorm(47)), type = 's', main = 'plot(x, type = 's')')points(x, cex = .5, col = 'dark red')# }Community examples
rdocumentationorg@mennovr.nl at Nov 17, 2020 graphics v3.6.2
```r # Plot with multiple lines in different color: plot(sin,-pi, 4*pi, col = 'red') plot(cos,-pi, 4*pi, col = 'blue', add = TRUE) ```
rdocumentationorg@mennovr.nl at Nov 17, 2020 graphics v3.6.2
```r ## Plot with multiple lines in different color: plot(sin,-pi, 4*pi, col = 'red') plot(cos,-pi, 4*pi, col = 'blue', add = TRUE) ```
plot(basedata1$iq, basedata$read_ab, main='Diagrama de Dispersión', xlab = 'read_ab', ylab = 'iq')
ltseiden@gmail.com at Dec 13, 2020 graphics v3.4.0
## Linear Regression ExamplePlot points and add linear regression model line:```rlinreg <- lm(dist ~ speed, cars)linreg_coeffs <- coef(linreg)lineq <- paste('distance = ', linreg_coeffs[2], ' * speed + ', linreg_coeffs[1])plot(cars, main = 'Car distance by speed', sub = lineq, xlab = 'speed', ylab = 'distance', pch = 19)abline(linreg, col = 'blue')```
richie@datacamp.com at Jan 17, 2017 graphics v3.3.2
Pass a numeric vector to the `x` and `y` arguments, and you get a scatter plot. The `main` argument provides a [`title()`](https://www.rdocumentation.org/packages/graphics/topics/title). ```{r} plot(1:100, (1:100) ^ 2, main = 'plot(1:100, (1:100) ^ 2)') ``` If you only pass a single argument, it is interpreted as the `y` argument, and the `x` argument is the sequence from 1 to the length of `y`. ```{r} plot((1:100) ^ 2, main = 'plot((1:100) ^ 2)') ``` `cex` ('character expansion') controls the size of points. `lwd` controls the line width. `pch` controls the shape of points - you get 25 symbols to choose from, as well as alphabetic characters. `col` controls the color of the points. When `pch` is `21:25`, the points also get a background color which is set using `bg`. [`points()`](https://www.rdocumentation.org/packages/graphics/topics/points) for more on how to change the appearance of points in a scatter plot. ```{r} plot( 1:25, cex = 3, lwd = 3, pch = 1:25, col = rainbow(25), bg = c(rep(NA, 20), terrain.colors(5)), main = 'plot(1:25, pch = 1:25, ...)' ) ``` If you specify `type = 'l'`, you get a line plot instead. See [`plot.default()`](https://www.rdocumentation.org/packages/graphics/topics/plot.default) for a demonstration of all the possible values for type. ```{r} plot( (1:100) ^ 2, type = 'l', main = 'plot((1:100) ^ 2, type = 'l')' ) ``` `lty` controls the line type. `col` and `lwd` work in the same way as with points. [`lines()`](https://www.rdocumentation.org/packages/graphics/topics/lines) for more on how to change the appearance of lines in a line plot. ```{r} plot( (1:100) ^ 2, type = 'l', lty = 'dashed', lwd = 3, col = 'chocolate', main = 'plot((1:100) ^ 2, type = 'l', lty = 'dashed', ...)' ) ``` It is best practise to keep your `x` and `y` variables together, rather than as separate variables. ```{r} with( cars, plot(speed, dist, main = 'with(cars, plot(speed, dist))') ) ``` The formula interface, similar to modeling functions like [`lm()`](https://www.rdocumentation.org/packages/stats/topics/lm), makes this convenient. See [`plot.formula()`](https://www.rdocumentation.org/packages/graphics/topics/plot.formula) for more information. ```{r} plot( dist ~ speed, data = cars, main = 'plot(dist ~ speed, data = cars)' ) ``` If you pass a two column data frame or matrix then the columns are treated as the x and y values. So in this case, you can simply do: ```{r} plot(cars, main = 'plot(cars)') ``` The [`lines()`](https://www.rdocumentation.org/packages/graphics/topics/lines), [`points()`](https://www.rdocumentation.org/packages/graphics/topics/points) and [`title()`](https://www.rdocumentation.org/packages/graphics/topics/title) functions add lines, points and titles respectively to an existing plot. ```{r} plot(cars) lines(lowess(cars)) title('plot(cars); lines(lowess(cars))') ``` If the `x` variable is categorical, `plot()` knows to draw a box plot instead of a scatter plot. See [`boxplot()`](https://www.rdocumentation.org/packages/graphics/topics/boxplot) for more information on drawing those. ```{r} with( sleep, plot(group, extra, main = 'with(sleep, plot(group, extra))') ) ``` Again, the formula interface can be useful here. ```{r} plot(extra ~ group, sleep, main = 'plot(extra ~ group, sleep)') ``` Axis limits can be set using `xlim` and `ylim`. ```{r} plot( (1:100) ^ 2, xlim = c(-100, 200), ylim = c(2500, 7500), main = 'plot((1:100) ^ 2, xlim = c(-100, 200), ylim = c(2500, 7500))' ) ``` You can set log-scale axes using the `log` argument. ```{r} plot( exp(1:10), 2 ^ (1:10), main = 'plot(exp(1:10), 2 ^ (1:10))' ) plot( exp(1:10), 2 ^ (1:10), log = 'x', main = 'plot(exp(1:10), 2 ^ (1:10), log = 'x')' ) plot( exp(1:10), 2 ^ (1:10), log = 'y', main = 'plot(exp(1:10), 2 ^ (1:10), log = 'y')' ) plot( exp(1:10), 2 ^ (1:10), log = 'xy', main = 'plot(exp(1:10), 2 ^ (1:10), log = 'xy')' ) ``` If you pass a table of counts for a vector, `plot()` draws a simple histogram-like plot. See [`hist()`](https://www.rdocumentation.org/packages/graphics/topics/hist) for a more comprehensive histogram function. ```{r} plot( table(rpois(100, 5)), main = 'plot(table(rpois(100, 5)))' ) ``` For multi-dimensional tables, you get a mosaic plot. See [`mosaicplot()`](https://www.rdocumentation.org/packages/graphics/topics/mosaicplot) for more information. ```{r} plot( table(X = rpois(100, 5), Y = rbinom(100, 10, 0.75)), main = 'plot(table(X = rpois(100, 5), Y = rbinom(100, 10, 0.75)))' ) ``` You can also pass functions to plot. See [`curve()`](https://www.rdocumentation.org/packages/graphics/topics/curve) for more examples. ```{r} plot( sin, from = -pi, to = 2 * pi, main = 'plot(sin, from = -pi, to = 2 * pi)' ) ``` Use the axis function to give fine control over how the axes are created. See [`axis()`](https://www.rdocumentation.org/packages/graphics/topics/axis) and [`Axis()`](https://www.rdocumentation.org/packages/graphics/topics/Axis) for more info. ```{r} plot( sin, from = -pi, to = 2 * pi, axes = FALSE, main = 'plot(sin, axes = FALSE, ...); axis(1, ...); axis(2)' ) axis( 1, # bottom axis pi * (-1:2), c(expression(-pi), 0, expression(pi), expression(2 * pi)) ) axis(2) # left axis ``` Further graphical parameters can be set using [`par()`](https://www.rdocumentation.org/packages/graphics/topics/par). See [`with_par()`](https://www.rdocumentation.org/packages/withr/topics/with_par) for the best way to use `par()`. ```{r} old_pars <- par(las = 1) # horizontal axis labels plot((1:100) ^ 2, main = 'par(las = 1); plot((1:100) ^ 2)') par(old_pars) # reset parameters ```
API documentation
In this article, you will learn to use par() function to put multiple graphs in a single plot by passing graphical parameters mfrow and mfcol.
Sometimes we need to put two or more graphs in a single plot.
R par() function
We can put multiple graphs in a single plot by setting some graphical parameters with the help of par() function. R programming has a lot of graphical parameters which control the way our graphs are displayed.
The par() function helps us in setting or inquiring about these parameters. For example, you can look at all the parameters and their value by calling the function without any argument.
3 Plots In R Squared
You will see a long list of parameters and to know what each does you can check the help section ?par. Here we will focus on those which help us in creating subplots.
Graphical parameter mfrow can be used to specify the number of subplot we need.
It takes in a vector of form c(m, n) which divides the given plot into m*n array of subplots. For example, if we need to plot two graphs side by side, we would have m=1 and n=2. Following example illustrates this.
3d Plot In R
This same phenomenon can be achieved with the graphical parameter mfcol.
The only difference between the two is that, mfrow fills in the subplot region row wise while mfcol fills it column wise.
Same plot with the change par(mfcol = c(2, 2)) would look as follows. Note that only the ordering of the subplot is different.
More Precise Control
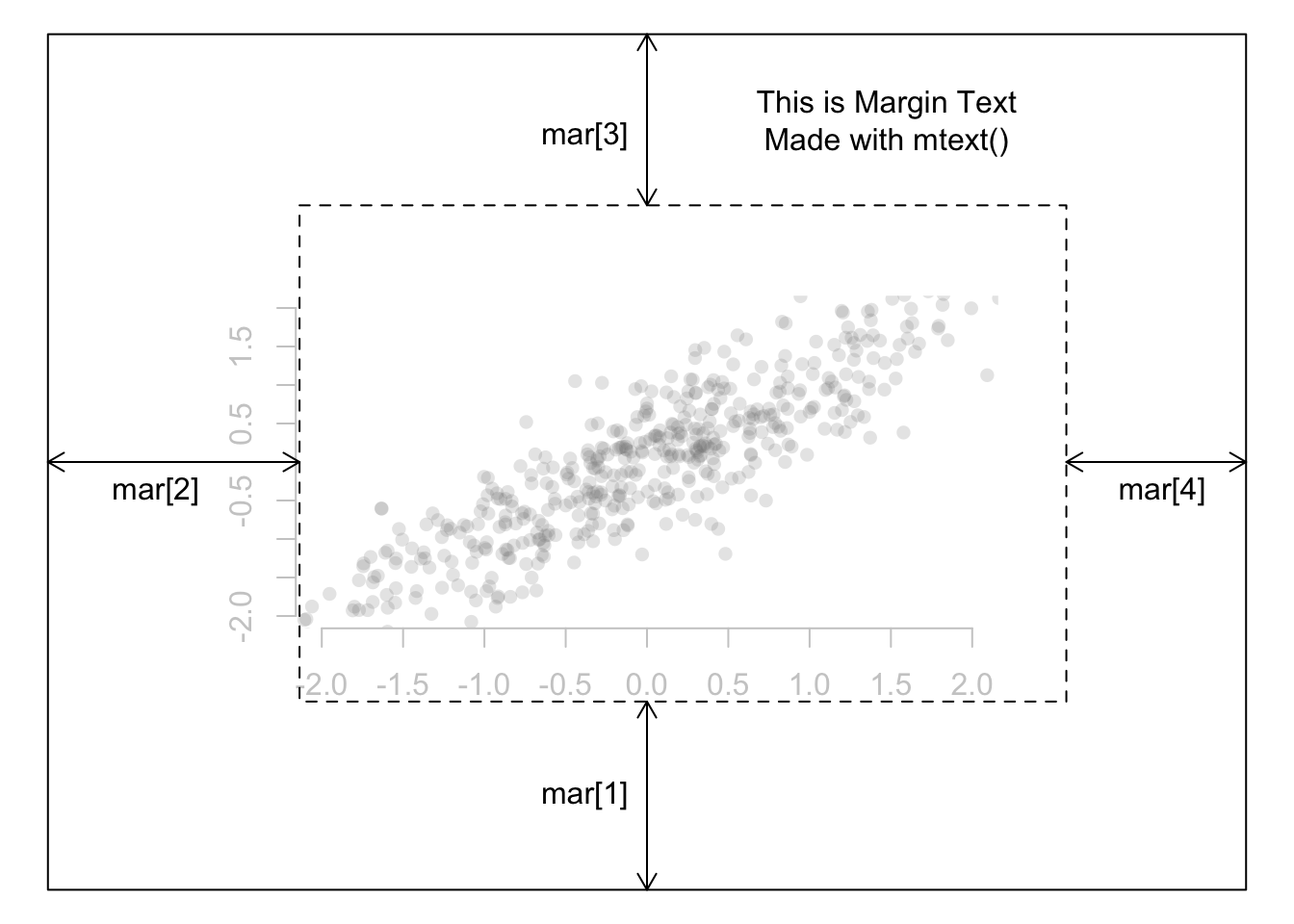
The graphical parameter fig lets us control the location of a figure precisely in a plot.
We need to provide the coordinates in a normalized form as c(x1, x2, y1, y2). For example, the whole plot area would be c(0, 1, 0, 1) with (x1, y1) = (0, 0) being the lower-left corner and (x2, y2) = (1, 1) being the upper-right corner.
Note: we have used parameters cex to decrease the size of labels and mai to define margins.
The numbers assigned to fig were arrived at with a hit-and-trial method to achieve the best looking plot.
- PREVIOUS
R Plot Function - NEXT
Saving a Plot in R